AI-powered and cloud-based Infection Control tool with Strain Typing (ST), and Antimicrobial Resistance Detection (AMR) profiling directly from MALDI-TOF MS ID files and FTIR data
A flexible and easy-to-use platform to take you beyond Microbial ID
(Research-Use-Only)
- Bacterial Sub-Species Typing
- Track outbreaks using ST/carbapenemase gene pairings representing widely distributed high-risk clones or clusters at a regional or hospital level. Enhance biosecurity and infection control in veterinary hospital environments.
- Co-create: work in consortia with different geographies by uploading your spectral libraries to a single cloud-based, secured location and then easily co-creating and sharing classification models to be used by your colleagues.
Dual Technology
The first software to combine MALDI-TOF MS and FTIR (psst, now also NGS) data in a single platform
Build your own prediction models
Easily create your training/validation sets and train with a complete variety of ML algorithms
Pre-trained Infection Control (clonal ST/AMR) models
Detect antimicrobial resistance in: K. pneumoniae, A. fumigatus, P. aeruginosa, C. difficile
Antibiotic Hydrolysis detection
Identify hydrolysys effect on Imipenem, Cefotaxime, Ceftriaxone and Meropenem
Biomarker analysis
Easy access to statistical tools: t-test, Pearson correlation, ROC curves and much more
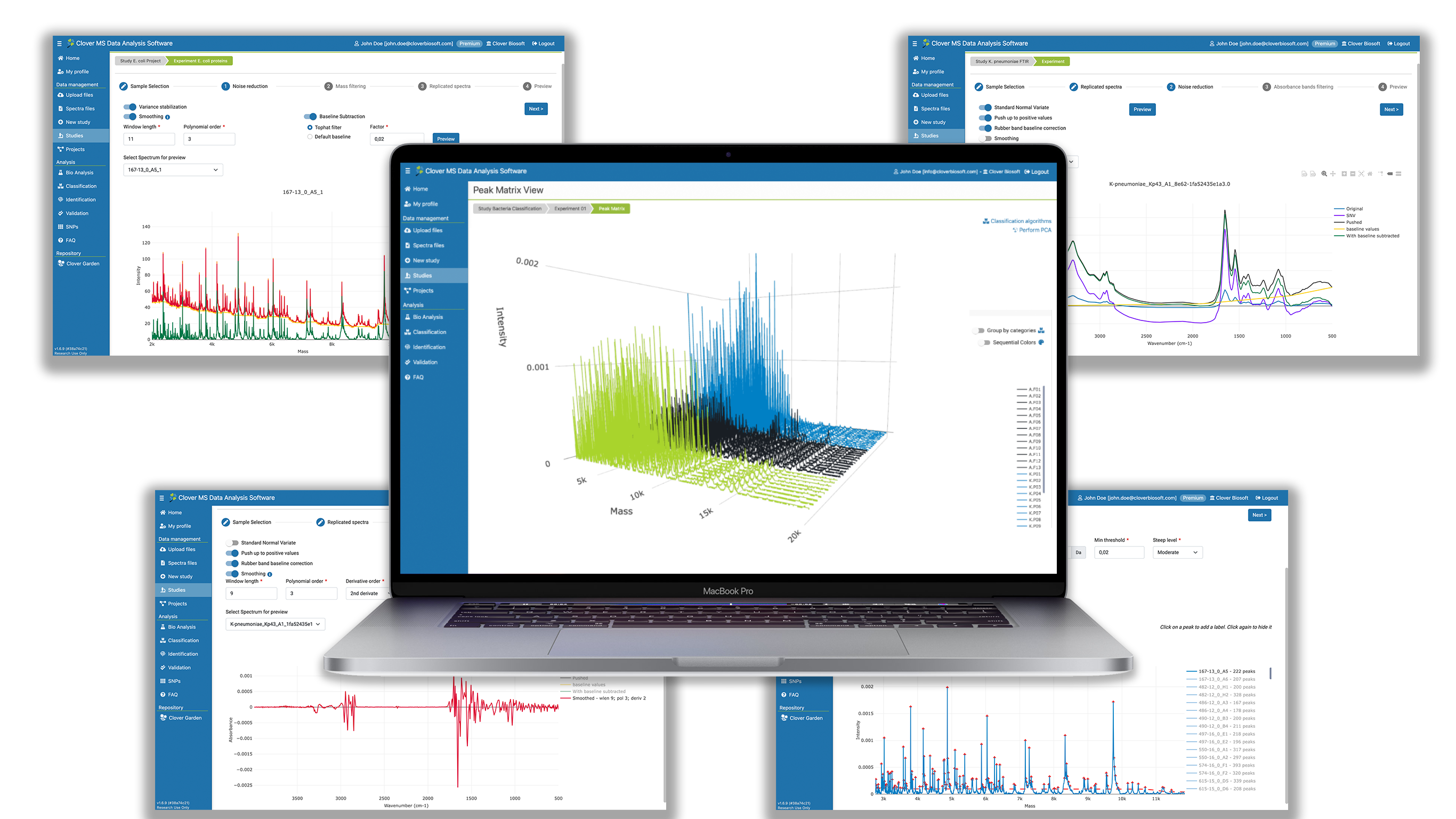
Quality control and reproducibility
Curate your data, discard low quality spectra and check for reproducibility in your experiments
How it works
Make the best use of the data you are already producing
Workflow 1: Subtyping or resistance marker detection using pretrained prediction models
Generate spectra with your MALDI or FTIR instrument
Generate spectra from any instrument and following the standard workflow already in use.
Upload to Clover MSDAS prediction tool
Just drop the spectra file(s) generated into one of our prediction models based on your microbial ID result.
Get your result in minutes
Follow the quick process and get your subtyping or AMR results in just a few minutes!
Workflow 2: Creating your own prediction models to solve any problem of your interest
Create a training dataset with your MALDI or FTIR spectra
Follow our assisted process to easily create and categorize your training, test and validation dataset.
Train your model
Choose from a wide range of Machine Learning algorithms. Use bioanalysis tools to take your knowledge further.
Validate your model
Save your trained model and use it later to blindly identify new samples from yours or a different lab.
All you can think of
Designed by microbiologists for microbiologists
AMR detection & subtyping
Pretrained prediction models to get more information on your Microbial ID-identified MALDI spectra.
- Identification of resistance mechanisms Carbapenemase Producing (CP) Klebsiella pneumoniae: KPC, OXA-48, NDM, VIM
- Azole-resistance detection in Aspergillus fumigatus complex isolates
- Prediction of high-risk clones of Pseudomonas aeruginosa: ST175 and ST235
- Prediction of Clostridioides difficile TER and ST181/ST027

Analyse
Analyze your spectra with specific tools from reproducibility to quality control checks.
- Biomarker Analysis with improved algorithms
- Outlier detection
- Spectra Quality Control
- Reproducibility test with Correlation analysis & Peak distribution
- Antibiotic Hydrolysis Detection Analysis
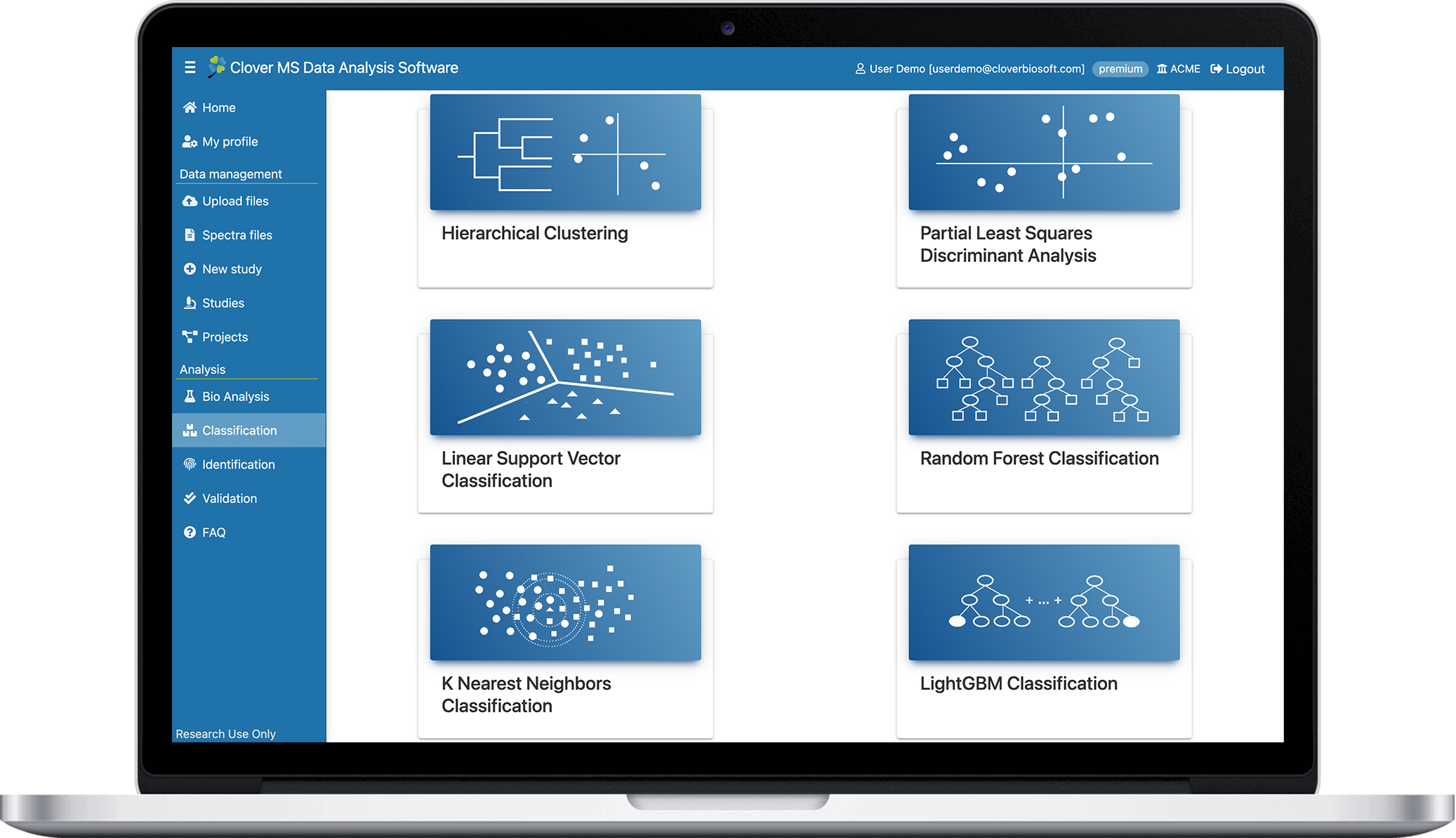
Train your model
- Best Machine Learning algorithms to make prediction models with your datasets
- Hierarchical clustering
- K-Means clustering
- Partial Least Squares Discriminant Analysis
- Linear Support Vector Classification
- Random Forest Classification
- K Nearest Neighbours Classification
- LightGBM Classification
- Clinic metrics, tables, plot and graphs from algorithms available for downloa
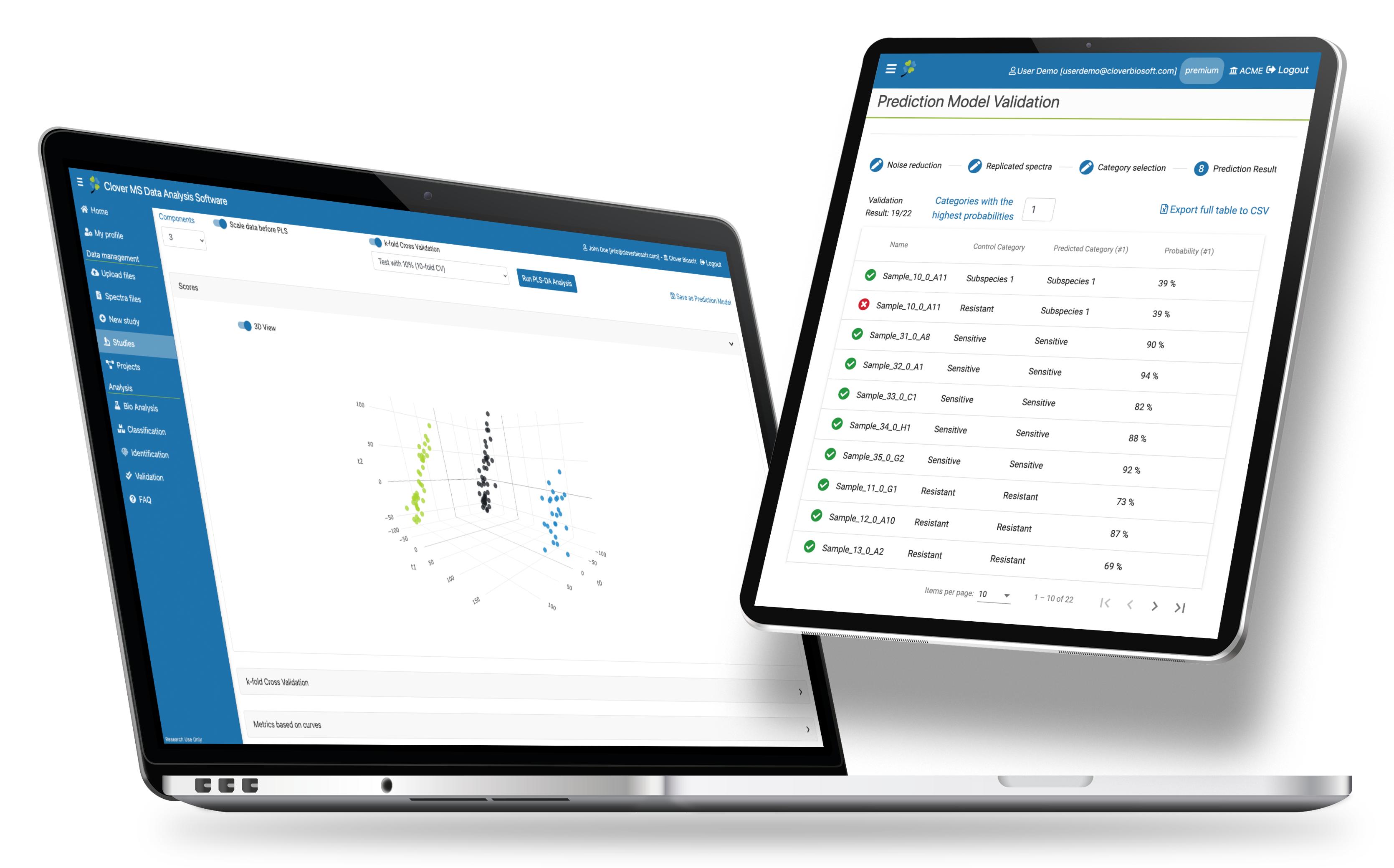
Predict & Identify
Validate your prediction models and use them to identify blind samples.
- Blind sample identification
- Validation of prediction models
- Multicentre validation
- Provide resistance score
"Your software literally saved our project"
– Dr. Belén Rodríguez, Hospital Gregorio Marañón, Spain
"This software is unique in its ability to process and compare MALDI and FTIR data together"
– Dr. Emmanuel Wey, Royal Free Hospital, UK
"The platform is flexible, informative and intuitive. Congratulations"
– Dr. Ângela Novais, UCIBIO, Portugal
"It’s just the workflow that we need. And sharing data with others is really important to us"
– Dr. Gianluca Foglietta, Ospedale Pediatrico Bambino Gesù, Italy
Choose the Right Plan for Your Microbial Analysis Needs
Lite Spectra Explorer
Limited functionality- Try it free for three months
- Uplad MALDI-TOF and FTIR data from any manufacturer
- 2 users per organisation
- Limited storage
- No access to extra modules
MALDI Masster
Premium MALDI account- Unlimited MALDI-TOF data storage
- MALDI-TOF MS full functionality
- 5 users per organisation
- Training course and email support
- 6 months and 3 months plan available
FTIR Prodigy
Premium FTIR account- Unlimited FTIR data storage
- FTIR full functionality
- 5 users per organisation
- Training course and email support
- 6 months and 3 months plan available
Spectra Maestro
Full access- Unlimited storage of any data type
- Full access to every module
- 10 users per organisation
- Training course and priority support
- 6 months and 3 months plan available
Coming soon!
New pay-per-use licenses to use our established classifiers for diagnostics of clonality, subtyping and prediction of antimicrobial resistance with blind samples. Stay tuned for more news!
Contact us
Ready To Get Started?
Phone Number
(+34) 630 961 713
Office
- Edif. Centro de Empresas PTS.
- Av. del Conocimiento, 41
- 18016 Granada, Spain
Copyright © 2024 I Privacy Policy
